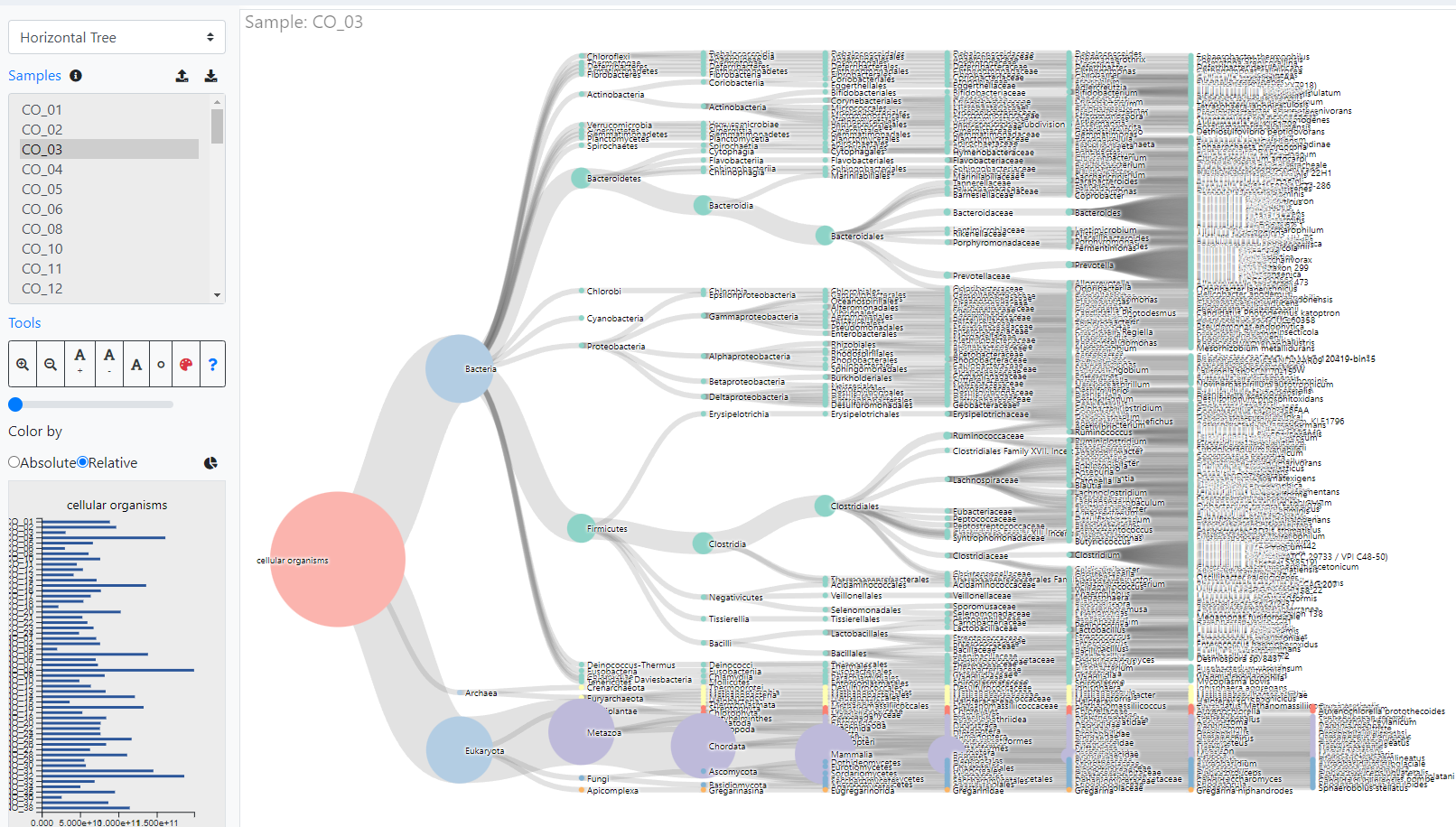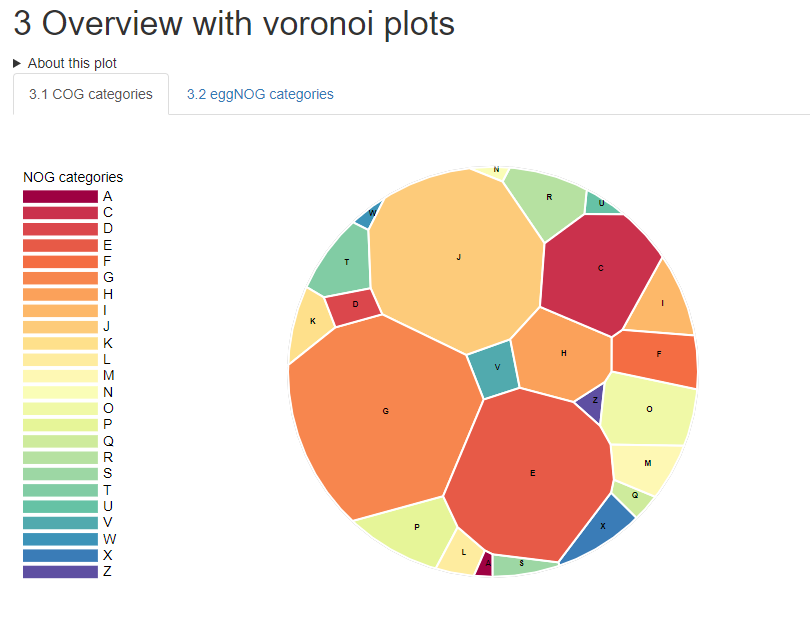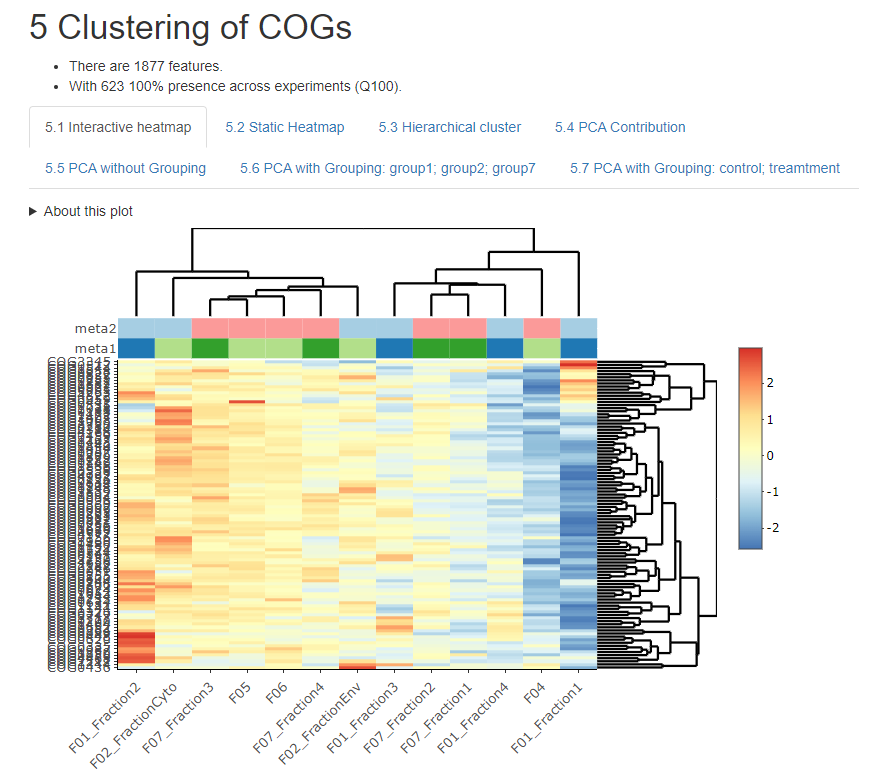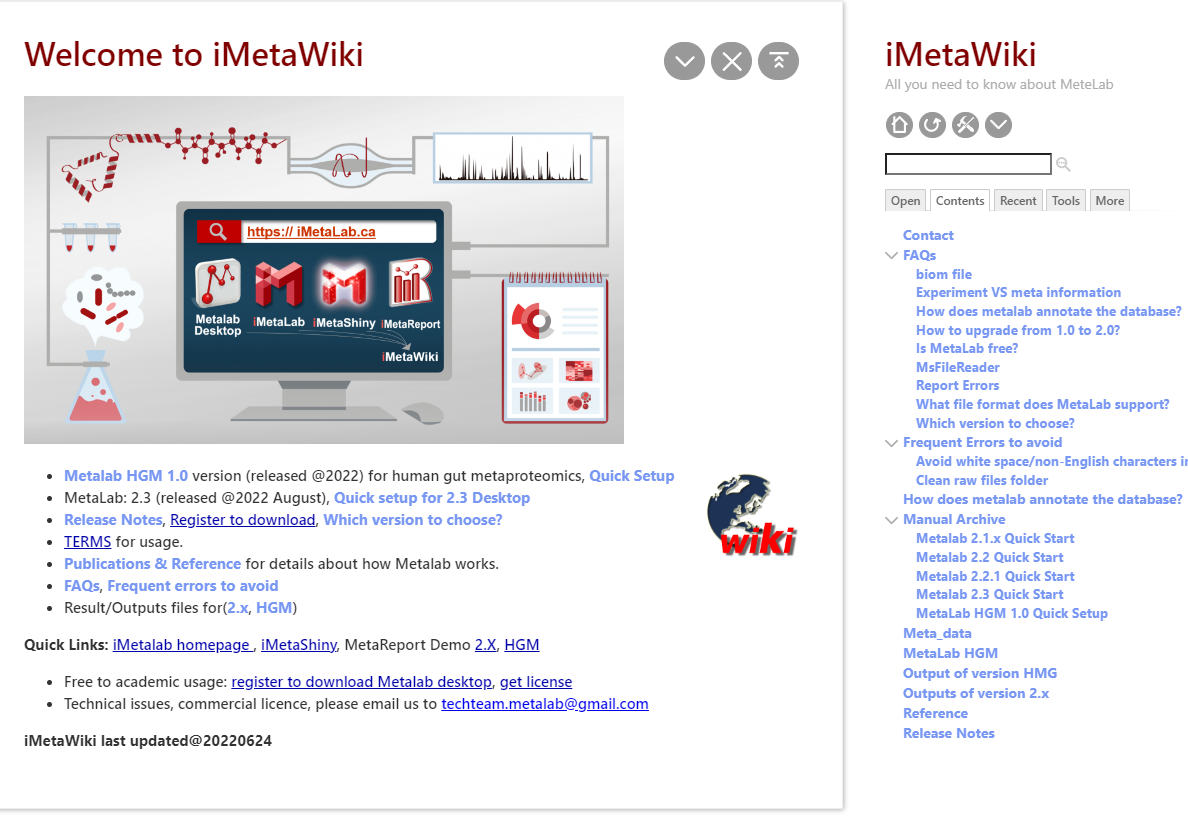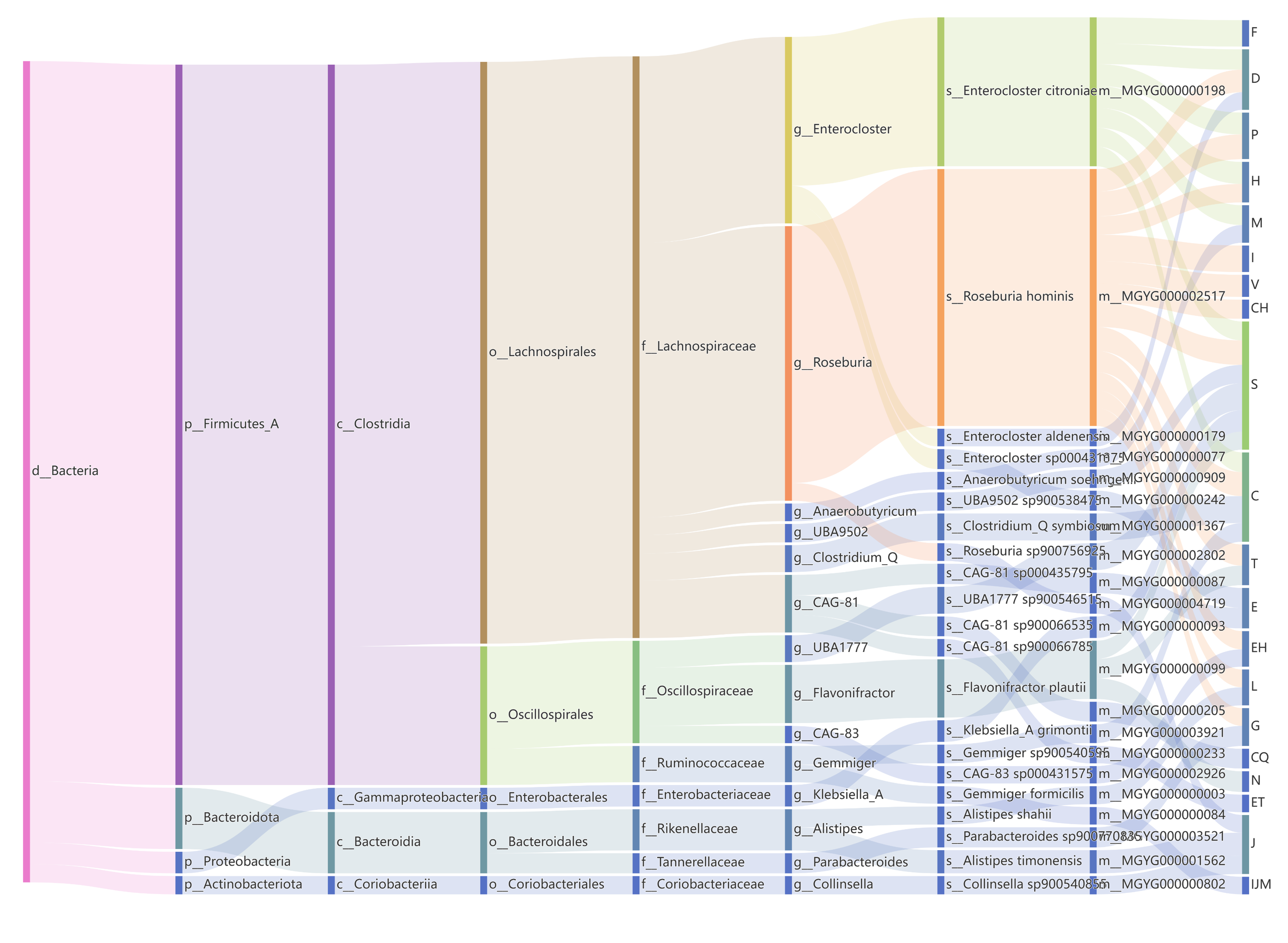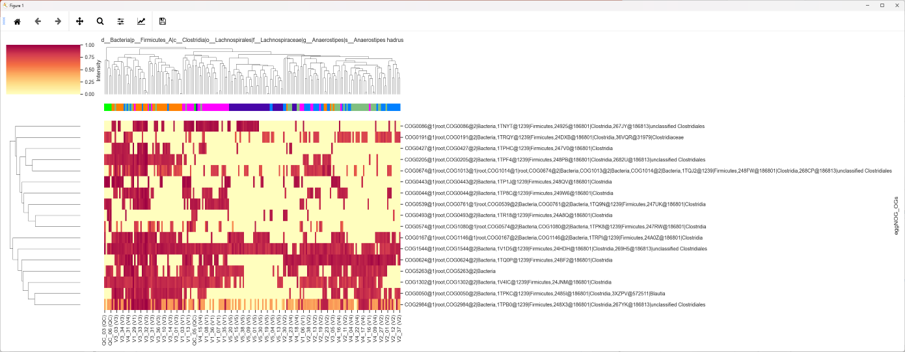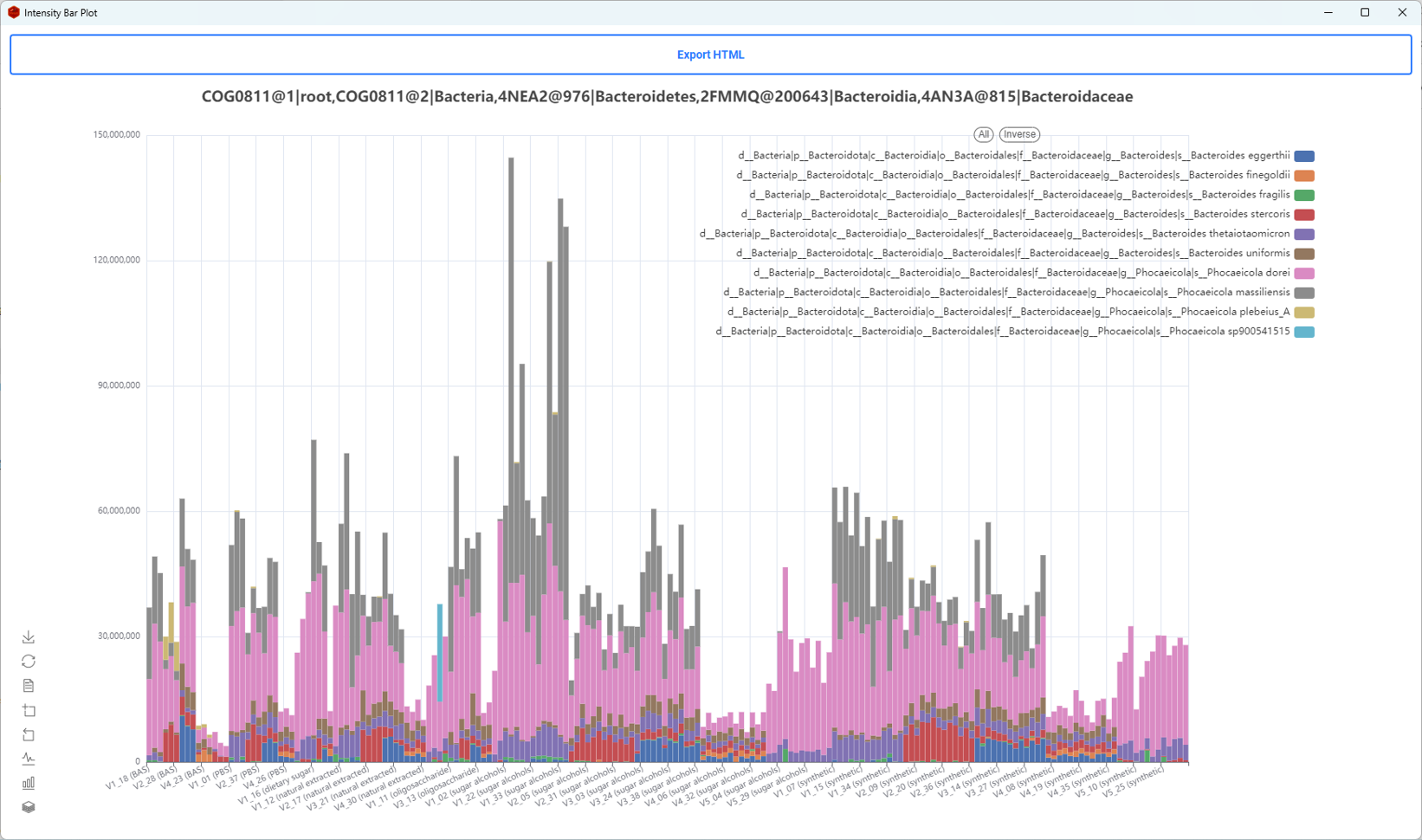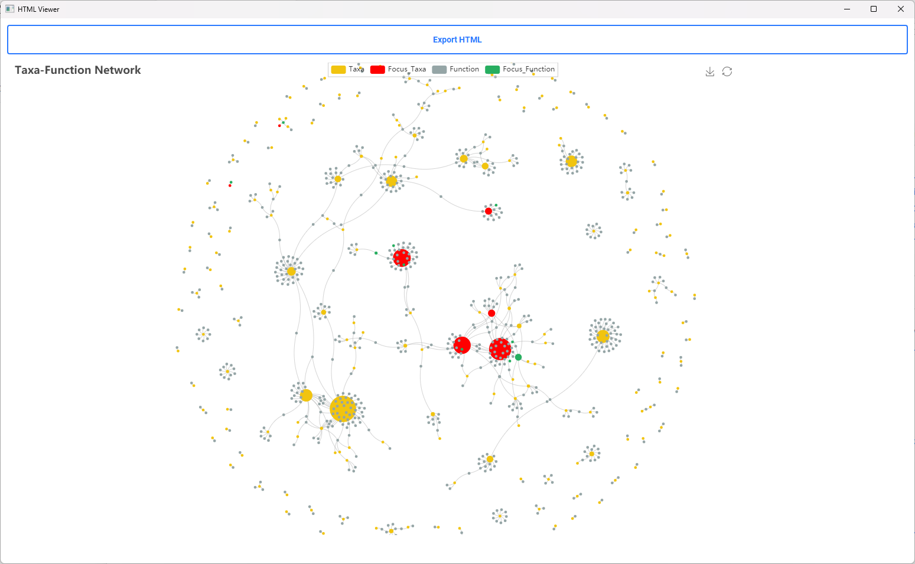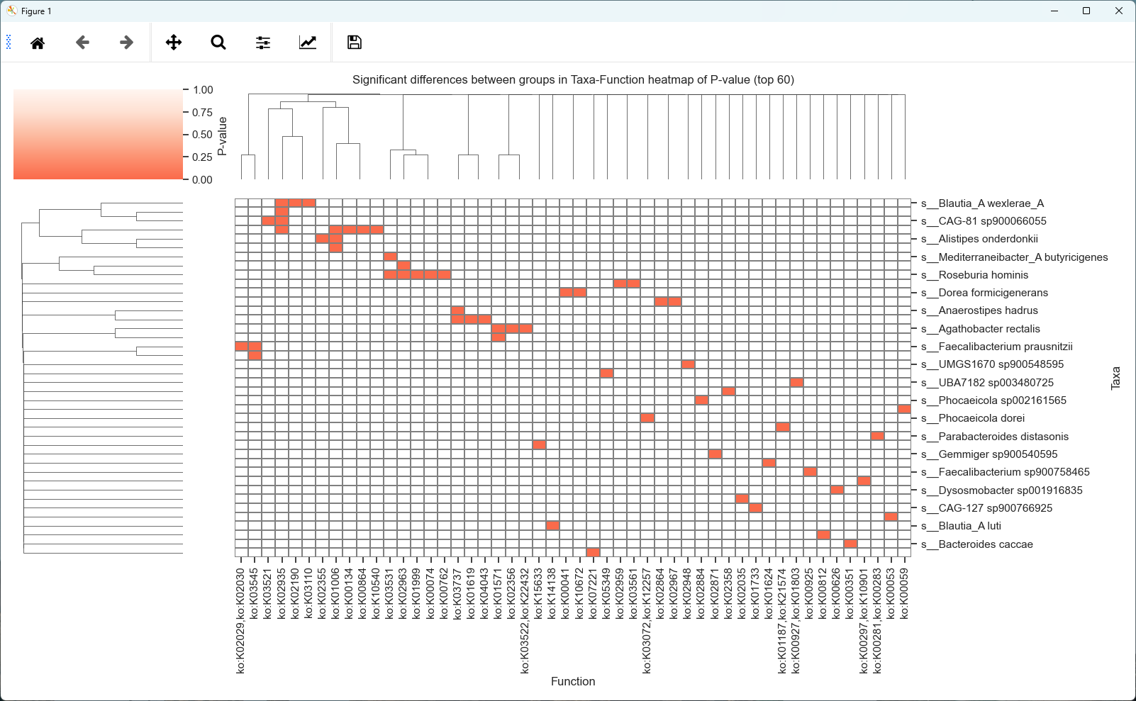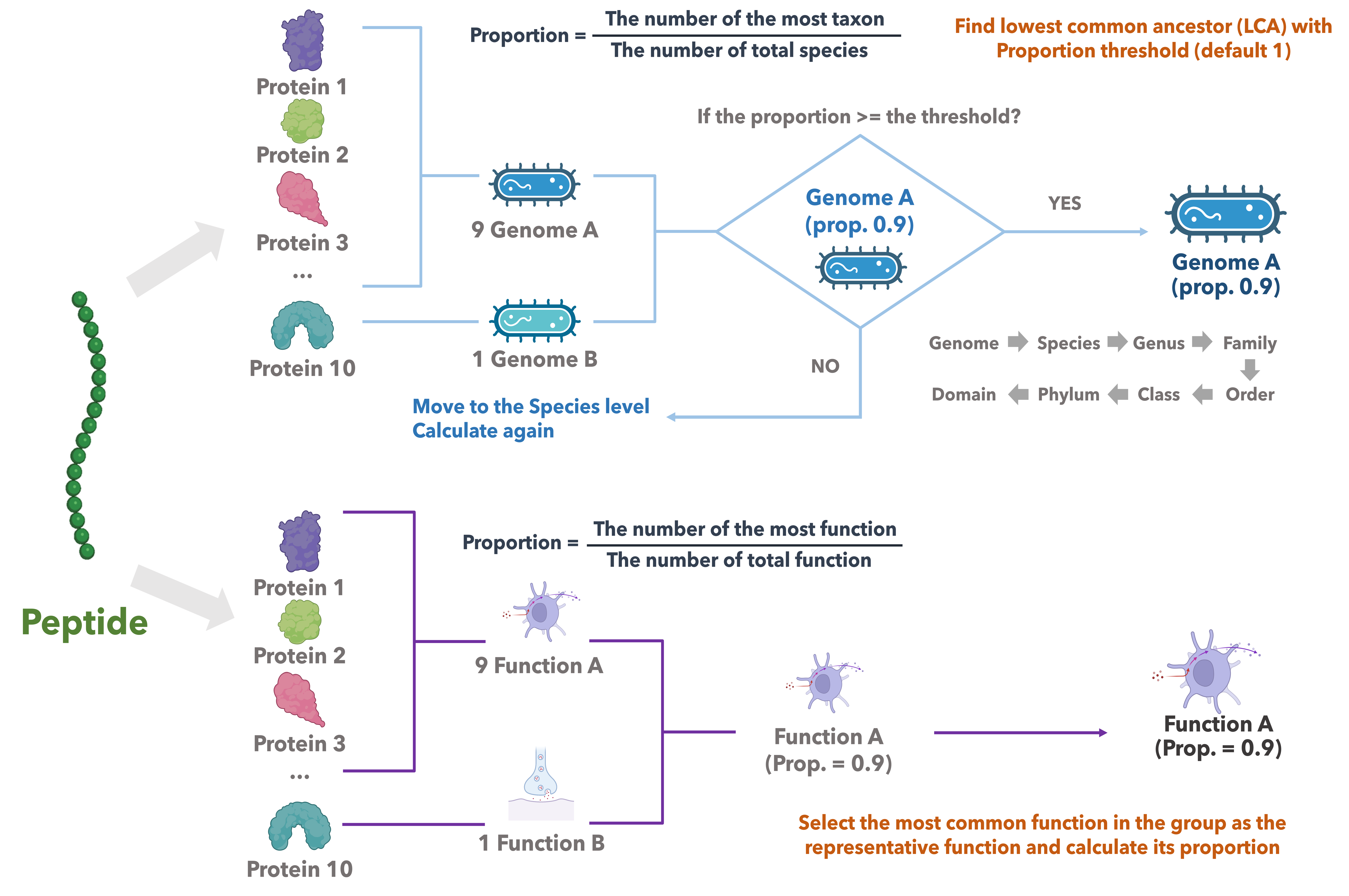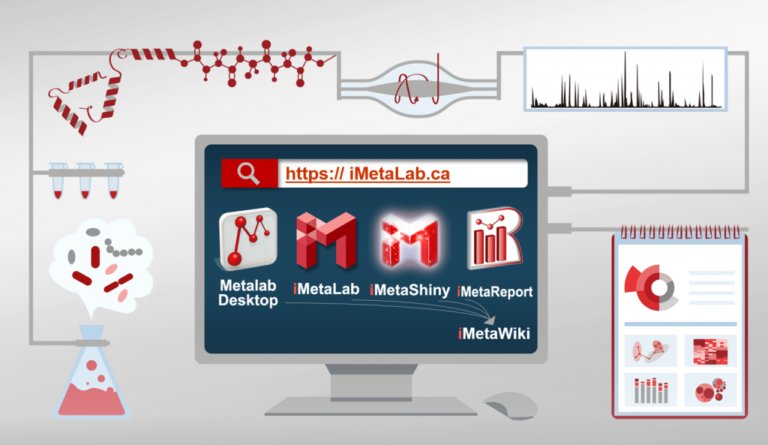
About iMetalab
- MetaLab was originally designed for human/mouse gut metaproteomics protein identification and quantification, rooted from the MetaPro-IQ workflow. Upon the rising request, we wrapped up the workflow into a desktop standalone version, with taxon and function analysis module built-in, and share the tool to the community for free.
- iMetalab represents the whole framework, including the Metalab Desktop version, automatic iMetaReport, iMetaShiny apps for data visualization and analysis, MetaX for taxon-function cross analysis. We aim to make iMetaLab platform a free and one-stop toolset for metaproteomics, with increasing amount of tools under active development. See publications about MetaLab
Evolution of Metalab suite

WorldWide Users
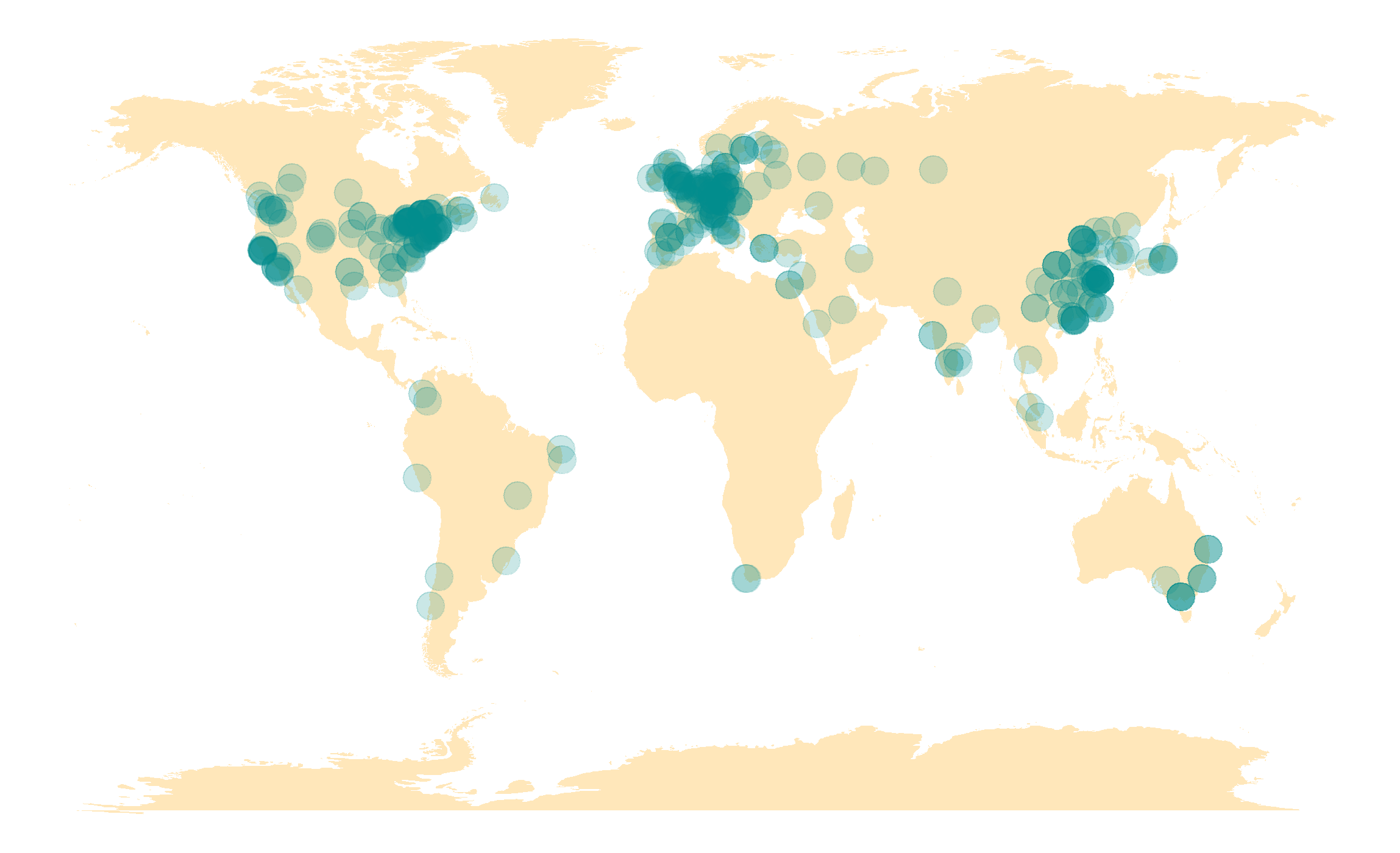
Why MetaLab? we are unique
Why Metalab? we are uniquE...
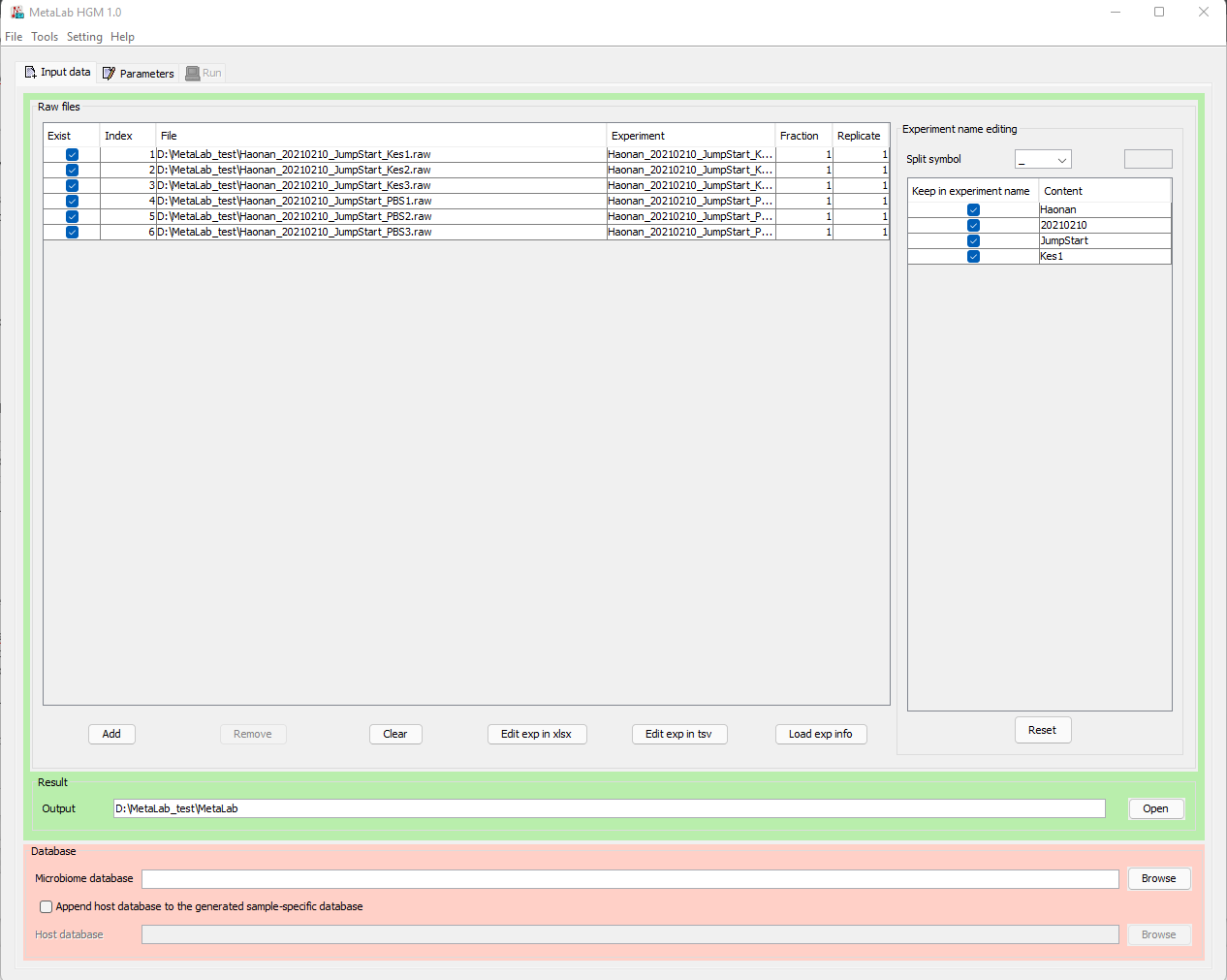
One stop toolkit for Protein Identification & Quantification for metaproteomic projects
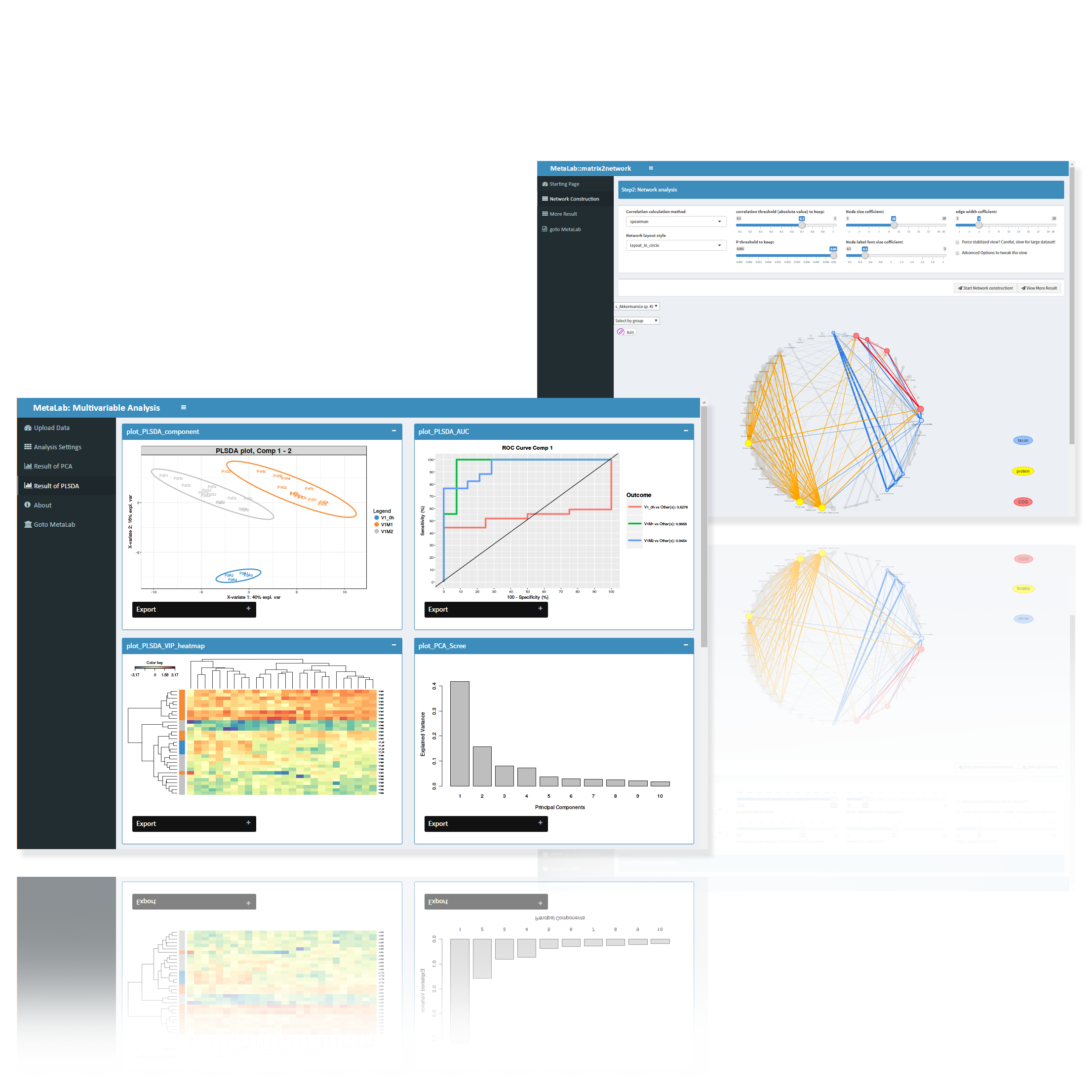
Automatic and detailed report on peptide, protein, taxon and functions
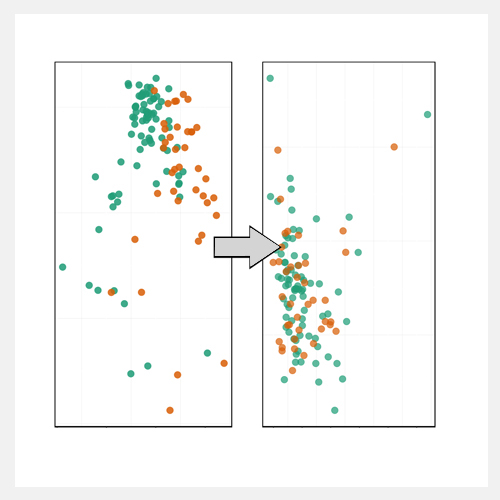
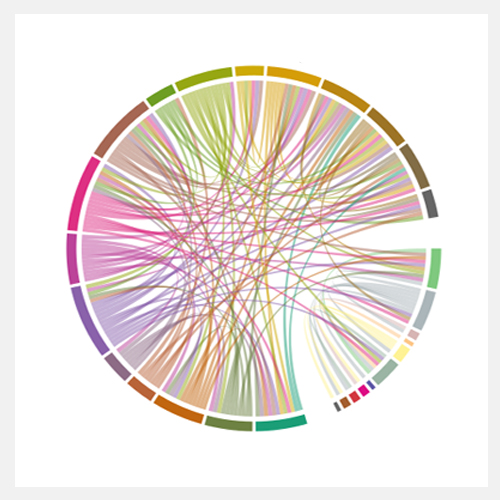
Indepth and comprehensive data analysis and publishable data visualization
Portfolio
Portfolio
- All
- iMetaReport
- MetaLab Desktop
- MetaX
- iMetaShiny
- MetaWiki
Team
Team
Multidisciplinary team works seamlessly

Daniel Figeys
Professor, School of Pharmaceutical Sciences University of OttawaCurrent Members
Kai Cheng (Ph.D.)
Metalab Desktop DeveloperZhibin Ning (Ph.D.)
Data analysis and visualizationQing Wu (Ph.D. Candidate)
MetaX developerZhongzhi Sun (Ph.D. Candidate)
MetaPep developerFormer Members
Leyuan Li (Ph.D.)
Developer for iMetaShiny and iMetaReportCaitlin Simopoulos (Ph.D.)
Developer for PepFunkXu Zhang (Ph.D.)
MetaProIQBo Liao (Ph.D.)
Full stack web developerThomas Li
Java script developer for MetaMapOmar Nasr
Java script developer for MetaMap
Accessibility
Accessibility
For Academic
Free
A licence still needed for activation
For business
contact us
Please contact for a commercial license via techteam.metalab@gmail.com
tryout!
tryout!
Contact
Contact
Feel free to contact us
Our Address
451 Smyth Road, Ottawa, ON, Canada, K1H 8M5
Email Us
techteam.metalab@gmail.com
Call Us
+1 613-562-5800